
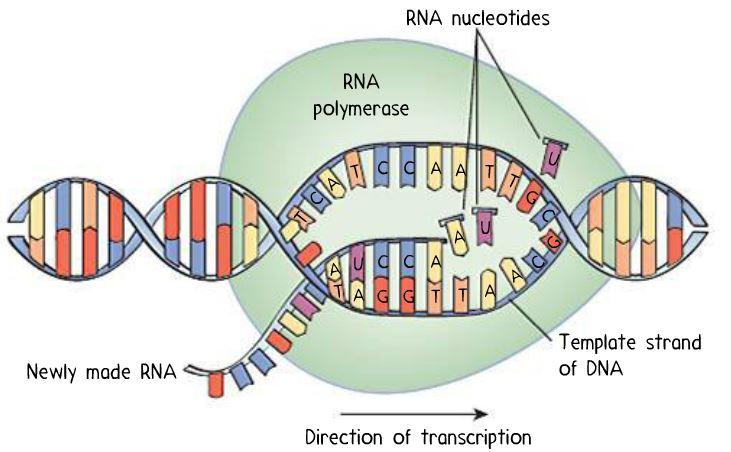
Helicase separates and begins unwinding dsDNA into single strands at the ori, forming two replication forks.Replication fork: Y-shaped region in the chromosome where both leading and lagging strands are replicated from the DNA template.Eukaryotes have numerous oris (e.g., 30,000–50,000 in humans) at which DNA replication begins simultaneously.Prokaryotes with circular genomes have a single origin.The origin of replication contains AT-rich sequences (e.g., TATA box regions), similar to those found in promoters.Specific proteins (comprising a prepriming complex) recognize and bind to the origin of replication.Origin of replication ( ori): a specific DNA sequence in the genome where DNA replication starts.The process of DNA replication Initiation Often up-regulated in tumor cells → excessive growth.N/A to bacteria and eukaryotes ( prokaryotes have circular chromosomes and do not require telomerase).Adds repeat sequences of DNA ( TTAGGG) to the 3′ end of linear chromosomes.A reverse transcriptase ( RNA-dependent DNA polymerase).Links newly synthesized DNA fragments ( Okazaki fragments) by catalyzing the formation of phosphodiester bonds.3′→5′ exonuclease activity for proofreading the synthesized strand.5′→3′ exonuclease activity for excising the RNA primer.Gaps between fragments are filled after primer removal.RNase H and FEN-1 ( flap endonuclease-1).RNase H and DNA polymerase I ( 5′→3′ exonuclease activity → excision of RNA primer ).Removal of primers (by replacing them with a DNA fragment).3′→5′ proofreading exonuclease activity.Extends the leading strand (until it reaches the preceding fragment's primer).DNA polymerase III: inhibited by certain drugs (e.g., antiretrovirals) via chain termination (the modified 3′-OH group of the drug prevents further elongation of the nucleotide chain).Extends RNA primers by adding 50–100 nucleotides.Primase activity as a component of DNA polymerase α.Topoisomerase II: i nhibited by chemotherapeutic agents etoposide and teniposide.Both enzymes are inhibited by certain antibiotics (e.g., quinolone, ciprofloxacin ).Topoisomerase II ( DNA gyrase ) and topoisomerase IV.Cleaves both DNA strands for larger structural alterations of DNA.Anti-Scl-70 antibodies (present in many individuals with scleroderma) recognize topoisomerase I as their antigen.Inhibited by chemotherapeutic agents irinotecan and topotecan.And has ligase activity to reseal the ligated strand.Has nuclease activity to cut DNA strands.Remove supercoils that develop during DNA elongation as parent DNA is separated by helicase.Prevents reannealing of separated strands.Single-stranded DNA-binding protein (SSBs) MCM (minichromosome maintenance) complex.Unwinds local segments of DNA double helix at replication fork.DNA repair mechanisms are, therefore, important to ensure genomic stability. Unrepaired DNA errors can cause mutations and/or cell destruction. External factors as well as internal cellular processes lead to alterations in the chemical structure of DNA. The process of DNA replication includes control mechanisms to keep the genetic information as stable as possible, but errors (e.g., the incorporation of a wrong base) still occur. This leads to the formation of two dsDNA molecules, each composed of one new and one original strand.
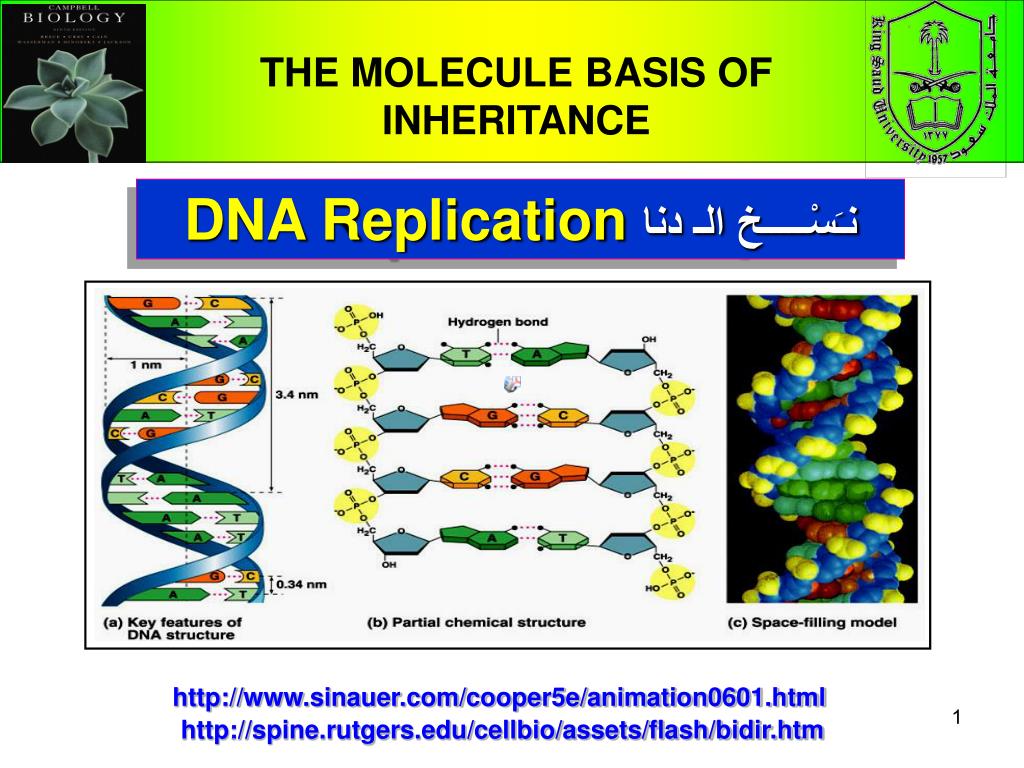
Specific proteins, including DNA polymerase, then synthesize a complementary daughter strand of double-stranded DNA (dsDNA) from each single strand template. Therefore, DNA replication requires that the DNA is loosened and the double helix is unwound. DNA is bound to proteins in the nucleus and is tightly packed. Cell division involves the duplication of a cell's entire DNA so that two genetically identical daughter cells arise from a single cell.


 0 kommentar(er)
0 kommentar(er)
